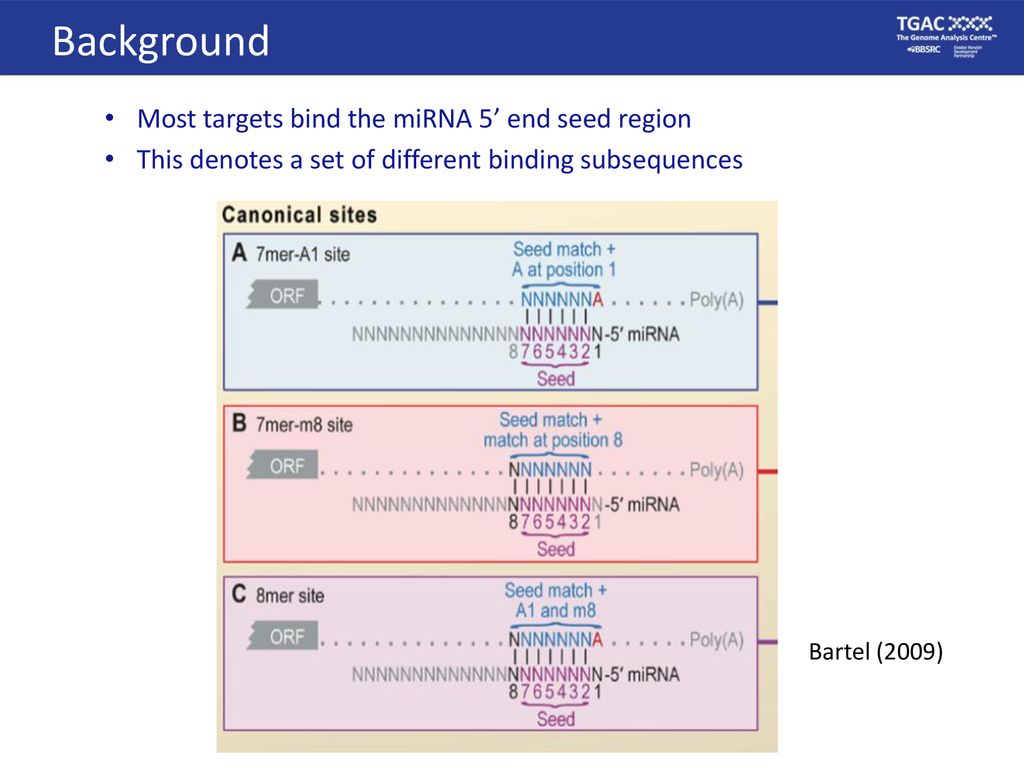
TargetScan is a target prediciton tool that predicts biological targets of miRNAs by searching for the presence of conserved 8mer and 7mer sites that match the seed region of each miRNA The target prediction software is frequently updated; PE, post eclosion PBM, post blood meal (D) Density plot of canonical miRNA seed matches in target mRNAs relative to the ligation site in all chimeras In the shuffled interactions (gray), each chimeric target region was randomly reassigned to a different miRNA (E) Predicted minimum free energy between miRNA and target mRNA found in chimeras The shuffled The nucleotides 28 in miRNA are termed 'seed,' which is the most crucial region for target recognition The seed region undergoes WatsonCrick base pairing with 3'UTR of mRNAs The degree of mRNA destabilization differs according to the class of the target site The different classes(of target sites to seed) are given below with increasing efficacy and preferential

Different Mirna Mrna Seed Site Interaction Patterns 6m Open I
Mirna seed region
Mirna seed region- Additionally, the degree of polysome occupancy of target mRNAs is dependent on the affinity between miRNA seed region and MRE, but not on miRNA abundance Moreover, at any given time the average proportion of polysomebound and unbound miRISC, called the polysome occupancy, was below 40% These data are in agreement with high rates of miRNAbound AGOFor the 1226 human miRNAs with SNPs in their seed region, 314 (256%) of them are located in miRNA clusters, whereas among the 1587 human miRNAs without SNPs in their seed region, only 3 (2%) of them are located in miRNA clusters (P = 606 × 10 −4, χ 2 test) (Table S2) miRNAs from the same cluster have the tendency to regulate the same sets of target genes and




Mapping The Human Mirna Interactome By Clash Reveals Frequent Noncanonical Binding Cell
Pasquinelli, 12) In contrast, most evidence indicates that miRNAs in land plants require more extensive pairing to their targets (Schwab et al, 05;Vealed that target hybridization to nucleotides of the seed region, at the 5′ end of an miRNA, was sufficient to induce moderate repression of expression In contrast, pairing to the 3′ region of the miRNA was not critical for silencing Our results suggest that the basepairing requirements for small RNAmediated repression in C reinhardtii are more similar to those of metazoans Partially complementary Watson–Crick base pairing of the miRNA seed region (nucleotides 2–8 from the 5′ end) to a target mRNA typically induces translational repression, while perfect base pairing of the miRNA results in siRNAinduced endonucleolytic cleavage that is mediated by AGO2 in vertebrates 30, 31 or by AGO1 and 10 along with other AGO paralogs in
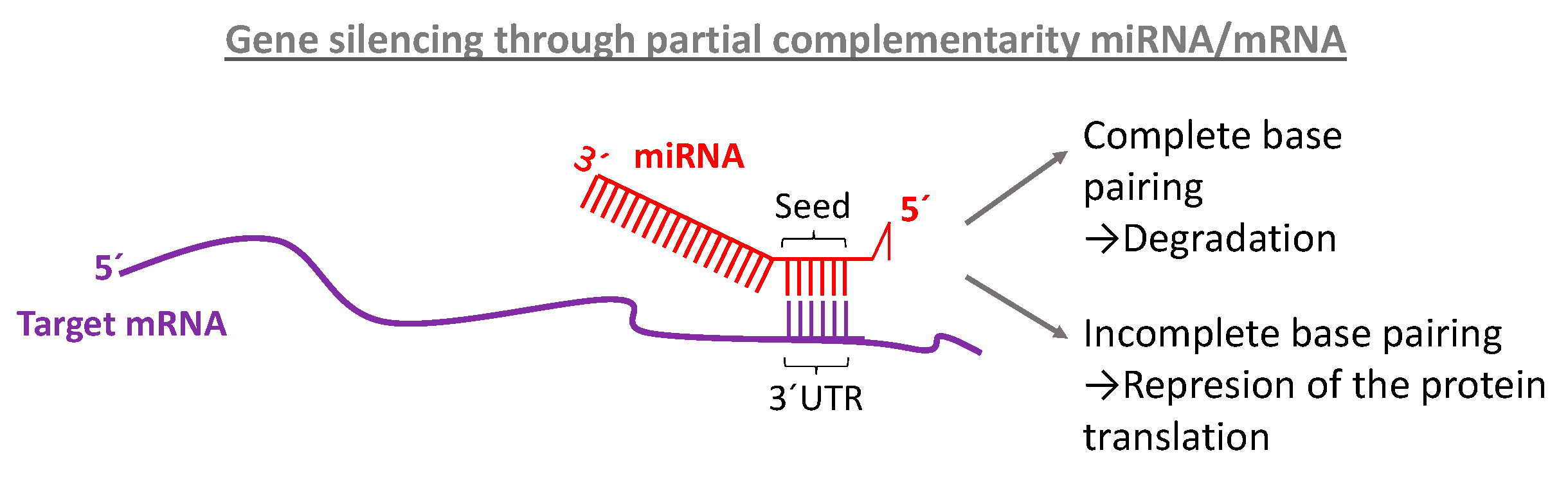
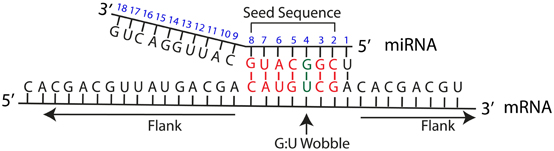
The seed sequence is essential for the binding of the miRNA to the mRNA The seed sequence or seed region is a conserved heptametrical sequence which is mostly situated at positions 27 from the miRNA 5´end Even though base pairing of miRNA and its target mRNA does not match perfect, the "seed sequence" has to be perfectly complementary Thus, the functions of miRNA 5′ terminal and seed regions in miRNAmediated gene silencing may differ Some miRNAs, such as let7 47,48, miR34 49, miR124 50 and miR125 49, are known to be The 5´UTR region of microRNAs is also known as seed region (nucleotides 1 through 8) and has the most crucial impact on targeting and function MicroRNAs do not require perfect complementarity for target recognition and a single microRNA is capable of regulating up to hundred or more mRNA species
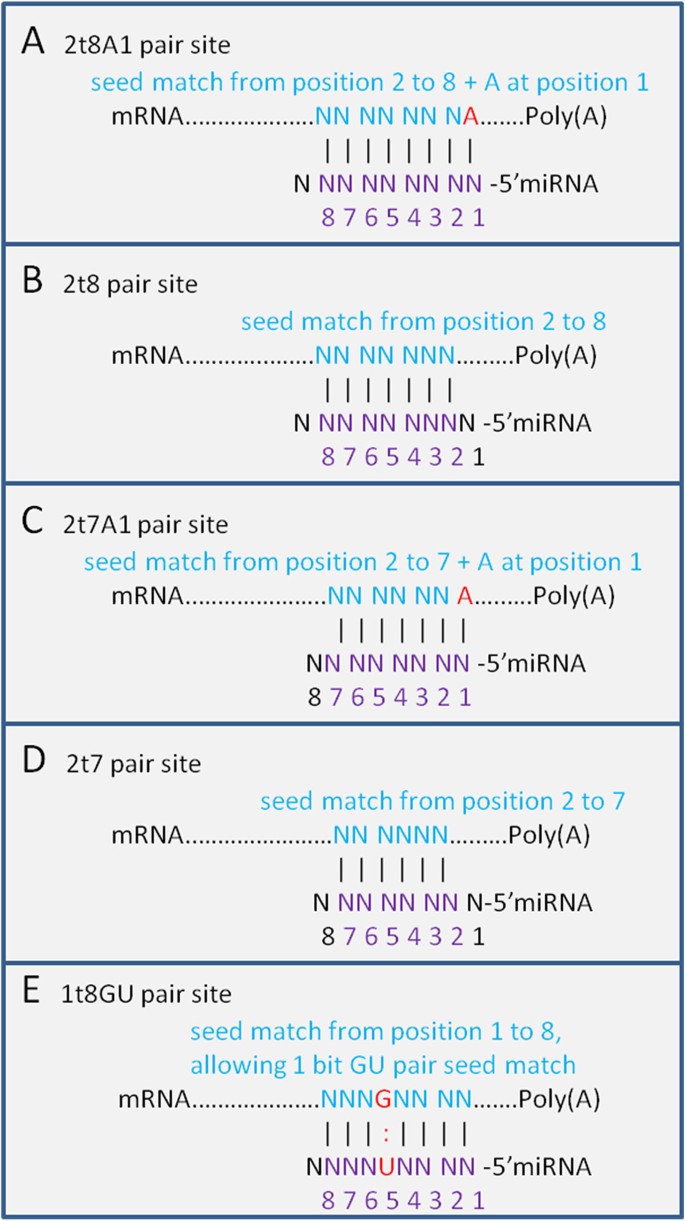
Download scientific diagram Different seed match regions of miRNAs miRnalyze follows a hierarchical pattern (8mer > 7merm8 > 7merA1 > 6mer) Many common features have been discovered for miRNA target regulation, such as perfect pairing of the miRNA 5′end (seed region) to the target site, as well as relatively low GC content of the target site, which results in increased site accessibility for miRNA binding 7,8,9,10,11,12,13 Despite steady progress in the field of miRNA target prediction, availableA mismatch tolerance test assay, based on pools of transgenic strains, revealed that target hybridization to nucleotides of the seed region, at the 5′ end of an miRNA, was sufficient to induce moderate repression of expression In contrast, pairing to the 3′ region of the miRNA was not critical for silencing Furthermore, we evaluated the silencing efficiencies of miRNAs with the




Microrna Single Nucleotide Polymorphisms And Diabetes Mellitus A Comprehensive Review Zhang 19 Clinical Genetics Wiley Online Library



Ijms Free Full Text Microrna Nanotherapeutics For Lung Targeting Insights Into Pulmonary Hypertension Html
Read Articles related to mirna seed region A range of mirna seed region information are available on echemicom A mismatch tolerance test assay, based on pools of transgenic strains, revealed that target hybridization to nucleotides of the seed region, at the 5′ end of an miRNA, was sufficient to induce moderate repression of expression In contrast, pairing to the 3′ region of the miRNA was not critical for silencing Our results suggest that the base‐pairing requirements for smallDevelop a scoring system (ie mRNA–miRNA partial complementarity, seed region, target position, sequence conservation features etc), which is then used to predict mRNA–miRNA interactions Each algorithm use slightly different scoring techniques, resulting in differences in prediction results A number of miRNA target prediction algorithms have been developed and tested for



Viral Mirna Adaptor Differentially Recruits Mirnas To Target Mrnas Through Alternative Base Pairing Elife



New Insights Into The Function Of Mammalian Argonaute2
When using default settings, TargetScan predicts biological targets of miRNAs by searching for the presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA (Lewis et al, 05) As options, predictions with only poorly conserved sites and predictions with nonconserved miRNAs are also provided Also identified are sites with mismatches in the seedTered miRNA seed regions with that of SNPs in nonclustered miRNA seed regions We found that the percent overlap of SNPs in clustered miRNA seed regions is much higher than that of SNPs in nonclustered miRNA seed regions (Student's ttest; An overview of the seedplussupplementarypaired structure reveals the miRNA and target RNA form two discontinuous duplexes one composed of the seed region, and a secondary duplex in the supplementary region (Fig 1C and D) Within the seed region, the target RNA pairs to nucleotides g2–g8, with the t1adenosine sequestered within the t1binding pocket, as reported



Small Rna Seq Reveals Novel Mirnas Shaping The Transcriptomic Identity Of Rat Brain Structures Life Science Alliance



Mir0c Microrna 0c
Here we show that point mutations in the seed region of miR96, a miRNA expressed in hair cells of the inner ear 8, result in autosomal dominant, progressive hearing loss This isThe members of a same miRNA may differ in the seed region and even one basepair difference between them can change their interaction with the target MicroRNAs (miRNAs) bind to mRNAs and target them for translational inhibition or transcriptional degradation It is thought that most miRNAmRNA interactions involve the seed region at the 5′ end of the miRNA The importance of seed sites is supported by experimental evidence, although there is growing interest in interactions mediated by the central region of the



Targetscan Non Canonical Sites




The Zswim8 Ubiquitin Ligase Mediates Target Directed Microrna Degradation
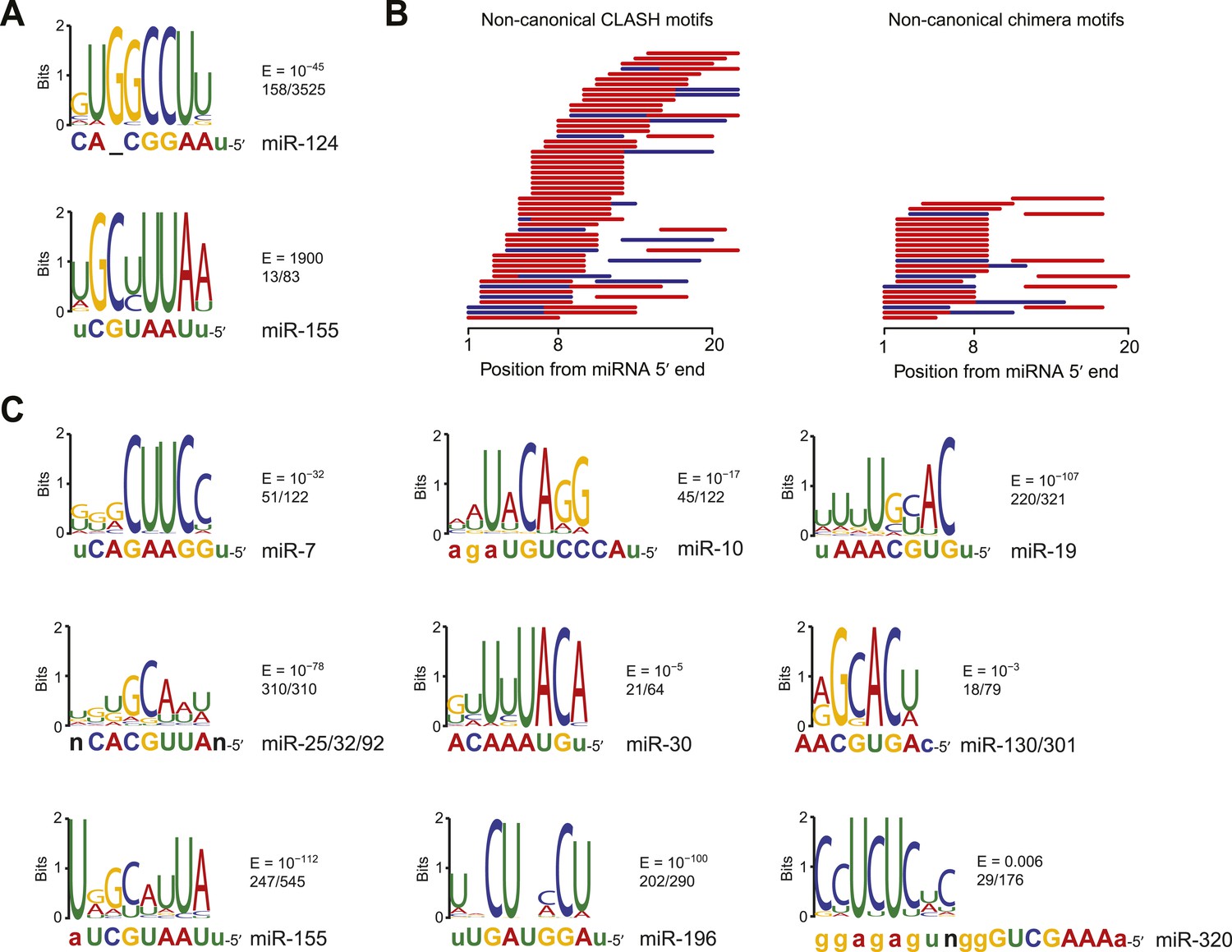
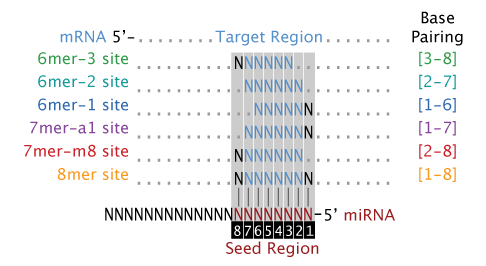
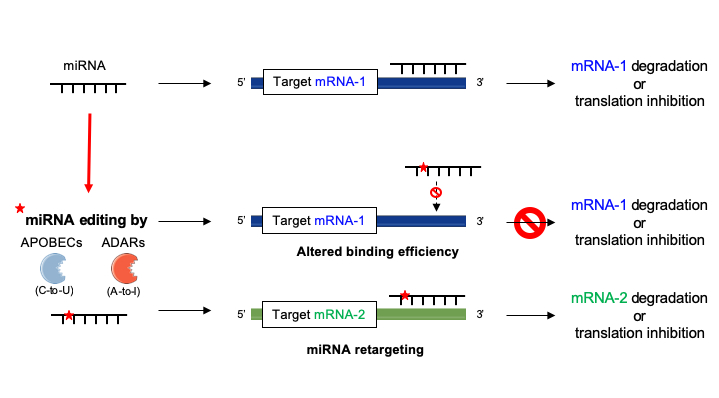
Base pairing of the miRNA seed region (positions 2–8 from the 5′ end of an miRNA) in animals is critical for target recognition and repression (Bartel, 09; Importance of seed and seedless matches Early studies on miRNA target recognition revealed nearperfect (contiguous) and conserved, WatsonCrick complementarity at the 5' miRNA end, which was called the "seed region" The seed is a 6–8 nt substring within the first 8 nucleotides, starting from the 5' miRNA end Typically, positions 2–7 from the 5' end areBy investigating its behavior in a dynamic context, we found that miRNA editing events in the seed region are not depended on miRNA expression, unprecedentedly providing insights on the targetome shifts derived from these modifications This reveals that miRNA editing acts under the influence of environmentally induced stimuliOur results show a miRNA editing activity trend



2




Mirna Mirna Targets Functional Analysis
The human genome encodes over 1000 miRNA genes that collectively target the vast majority of messenger RNAs (mRNAs) Basepairing of the socalled miRNA "seed" region with mRNAs identifies many thousands of putative targets Evaluating the strength of the resulting mRNA repression remains challenging, but is essential for a biologically informative ranking of potentialAs a result, we identified 48 SNPs in human miRNA seed regions and thousands of SNPs in 3' untranslated regions with the potential to either disturb or create miRNAtarget interactions Furthermore, we experimentally confirmed seven lossoffunction SNPs and one gainoffunction SNP by luciferase assay This is the first case of experimental validation of an SNP in an miRNAPositions 13–16 of the miRNA may aid in pairing as well , The miR195like sequence we have identified in #GU contains both the seed region and positions 13–16 of hsamiR195 and is 100% conserved in these regions It has also been




Got Target Computational Methods For Microrna Target Prediction And Their Extension Abstract Europe Pmc




Chemical Modifications In The Seed Region Of Mirnas 221 222 Increase The Silencing Performances In Gastrointestinal Stromal Tumor Cells Sciencedirect
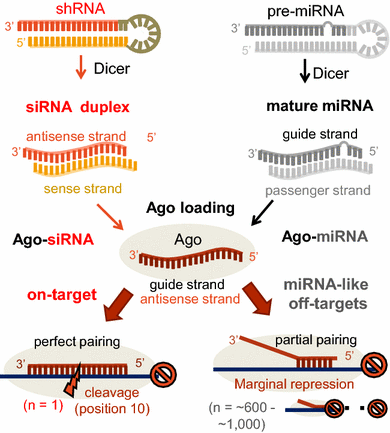
Clearly, the seed sequences of miRNA (identified with miRNA databases) should be avoided in siRNA design 69 In addition, the siRNA should have a low thermodynamic stability of the duplex between the seed region of the guide strand of siRNA and its target mRNA, since a low seedtarget duplex stability reduces the capability of siRNA to induce seeddependent off We identified 5252 miRNArelated SNPs potentially involving mastitisrelated traits We furtherly validated a candidate functional SNP (rs) in the seed region of btamiR29, which is associated with mastitis by affecting the interaction of btamiR29 and SPI1 in Chinese Holsteins This research is promising, as it may represent anTargetScan predicts biological targets of miRNAs by searching for the presence of 8mer, 7mer, and 6mer sites that match the seed region of each miRNA As an option, only conserved sites are predicted Also identified are sites with mismatches in the seed region that are compensated by conserved 3' pairing and centered sites In mammals, predictions are ranked based on the




Mirna Sequencing Report
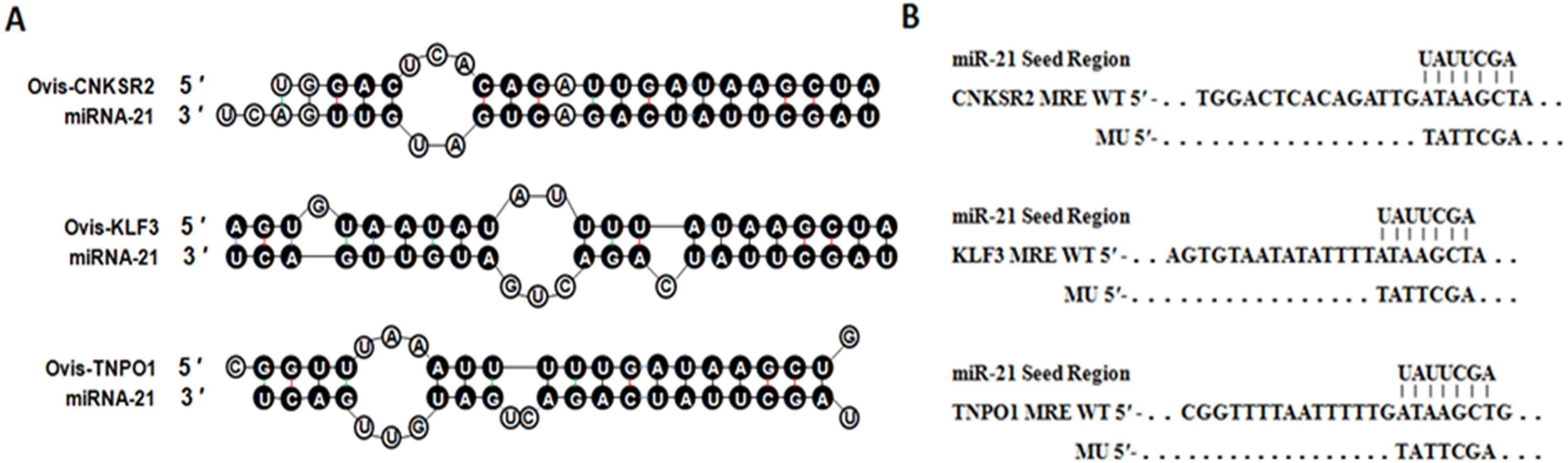



Identification Of Microrna 21 Target Genes Associated With Hair Follicle Development In Sheep Peerj
The recognition by the miRNA seed region of a perfectly matching sequence within a noncoding promoter transcript would recruit a nuclear RISC and induce cleavage of the antisense noncoding transcript, removal of chromatin effectors and derepression of the corresponding sense transcription 96,97 (Figure 1 A) An alternative mechanism is that the miRNA seed sequence A miRNA can target multiple mRNAs by functioning as a sequencespecific guide The binding is primarily directed through the interaction between the 5' ends of a miRNA, referred to as the "seed region," and the complementary 3' untranslated region (UTRs) of a target mRNA The nucleotides that would pair with the seed region of the miRNA are indicated by shading Full size image For all three miRNAs tested, mismatches causing greatest interference with inhibitor activity are located within positions corresponding to the seed region of the mature miRNAs (positions 3 to 8) and a second region closer to the 3' end (positions 13 to 18) (Figure




Canonical And Non Canonical Mirna Target Site Types A Diagram Of The Download Scientific Diagram




Microrna Regulation And Cardiac Calcium Signaling Circulation Research
Noncanonical sites are sites of interaction between a mRNA and a miRNA that do not involve the canonical pairing in the seed region (positions 2 7) of the miRNA They are of two types A 3' compensatory site is one in which strong 3' pairing (consequential miRNAtarget complementarity outside the seed region) compensates for an imperfect seed match (Friedman et al, 09) AThe latest version of this resource was released in August 15 miRSearchSecond, miRNA recognizes target mRNA with seedcomplementary sequences 2,3,32 The stability between seed region and target mRNA is a determinant of the efficacy of siRNA offtarget effects 33 Thus, the high stability of the seedtarget duplex might function as a positive factor, but that in the miRNA duplex the 5′ end might be a negative
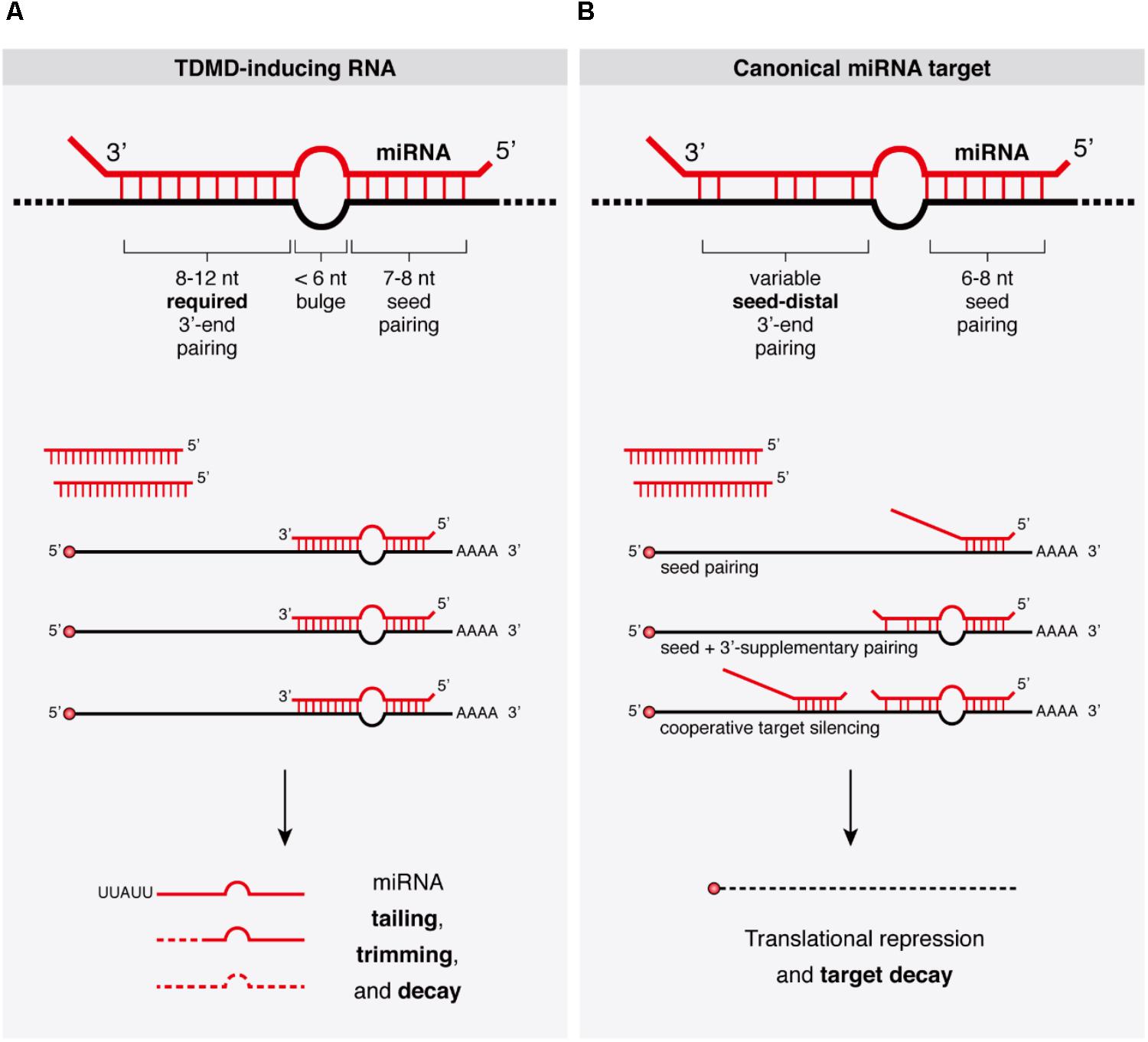



Frontiers Target Rnas Strike Back On Micrornas Genetics




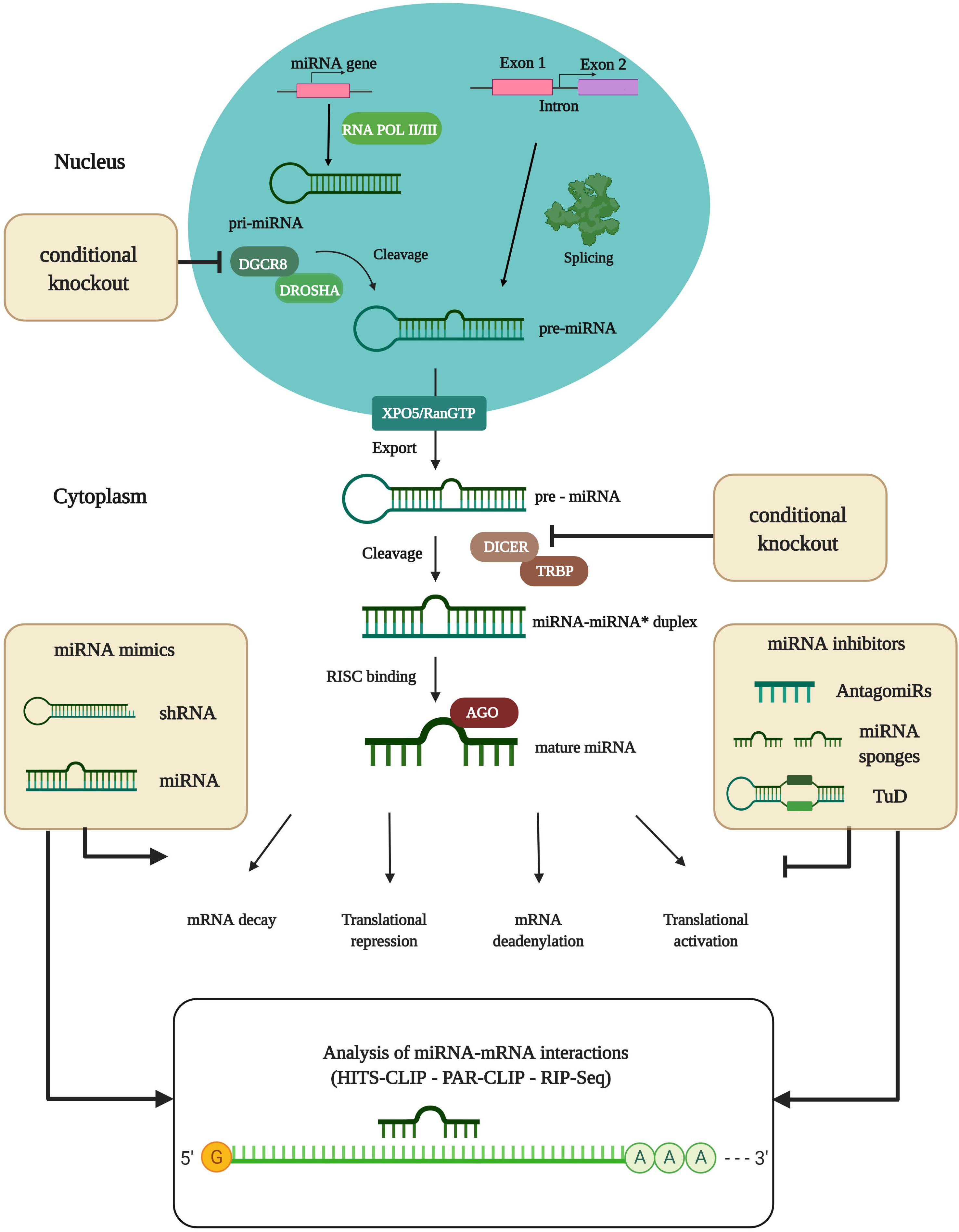
Mirna Introduction Biogenesis Nomenclature And Experimental Workflow
The function of miRNA is depending on the complementarity of the seed region of mature miRNA to the 3'UTR of the target mRNA gene, either undergoing mRNA cleavage or translational repression Strategies for miRNAbased therapies improving miRNA in disease can be achieved by the following approaches (a) Small molecule miRNA inhibitors can regulate miRNA Further, the potential action of a miRNA is mostly dependent on base pairing between the miRNA seed sequence and its target; The miRNA sequence can be separated into five functional domains that affect miRNAtarget recognition 5′ anchor (nt 1), seed sequence (nts 2–8), central region (nts 9–12), 3′ supplementary region (nts 13–16), and 3′ tail (nts 17–22) (Wee et al, 12) We anticipated that complementarity to the seed sequence of the cognate miRNA would be a prominent feature in
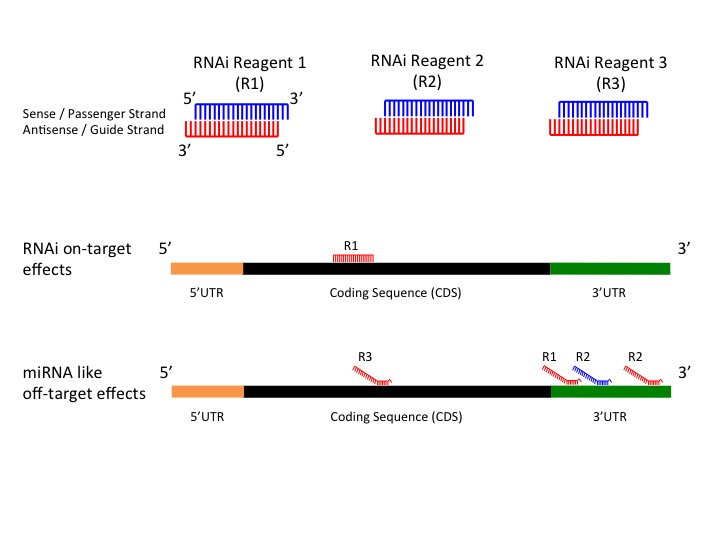



Gess




Correlation Between Mirna Targeted Gene Promoter F1000research
This shift was further demonstrated by a change in the seed binding regions of miR1243p, the most abundant retinal AGO2bound miRNA, and has known roles in regulating retinal inflammation Additionally, photoreceptor cluster miR1/96/1 were all among the most highly abundant miRNA bound to AGO2 Following damage, AGO2 expression was localized to the inner The miRNA seed region, positions 2–8 of the mature miRNAs, is particularly important for miRNA target recognition (Lewis et al 05) Many microRNAs are highly conserved across species (Bartel 04) The seed regions are more conserved than the flanking regions, indicating their functional importance (Wheeler et al 09) Consistently, the seed regions
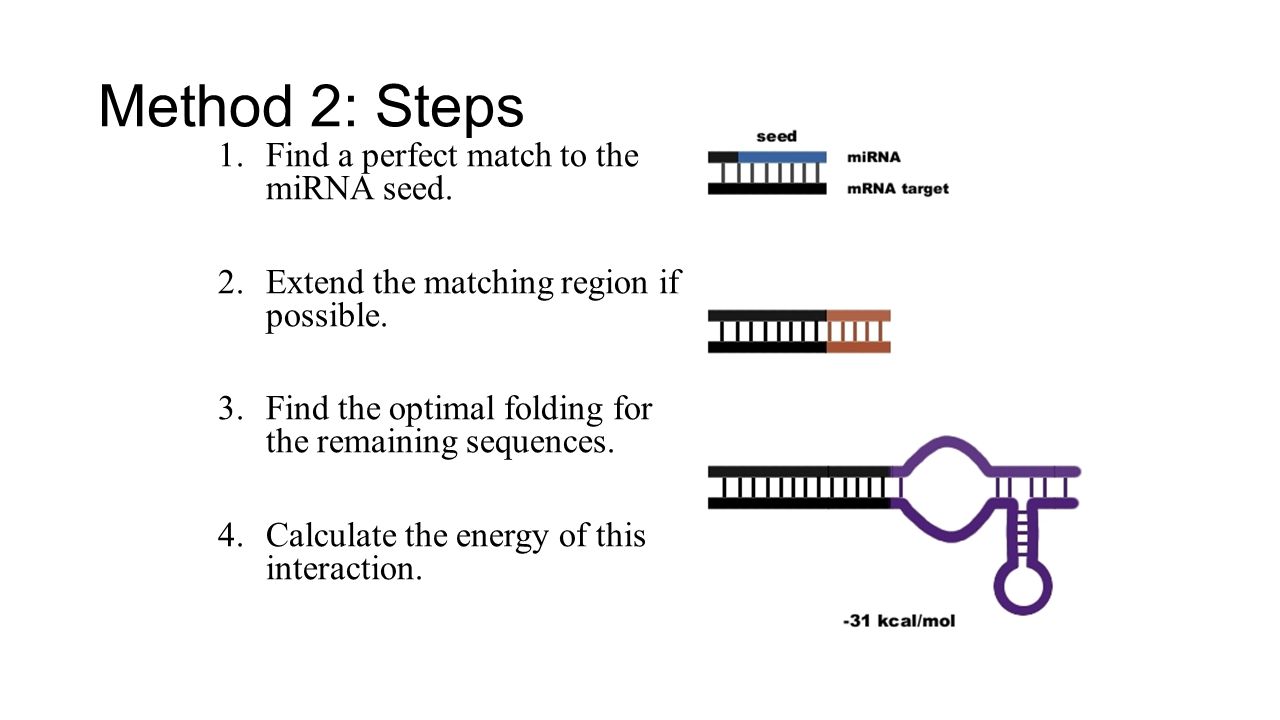



Microrna Computational Prediction And Analysis Ppt Download
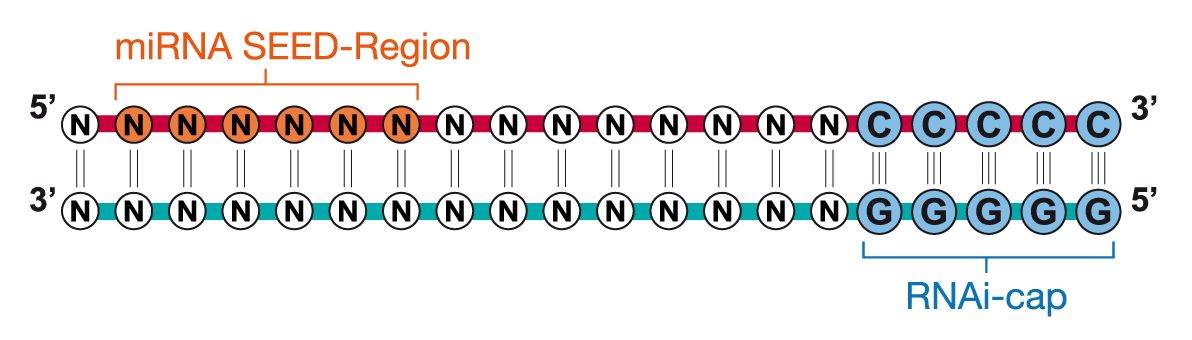



Riboxx Rna Technologies Benefits Of Rnai Cap For Mirna




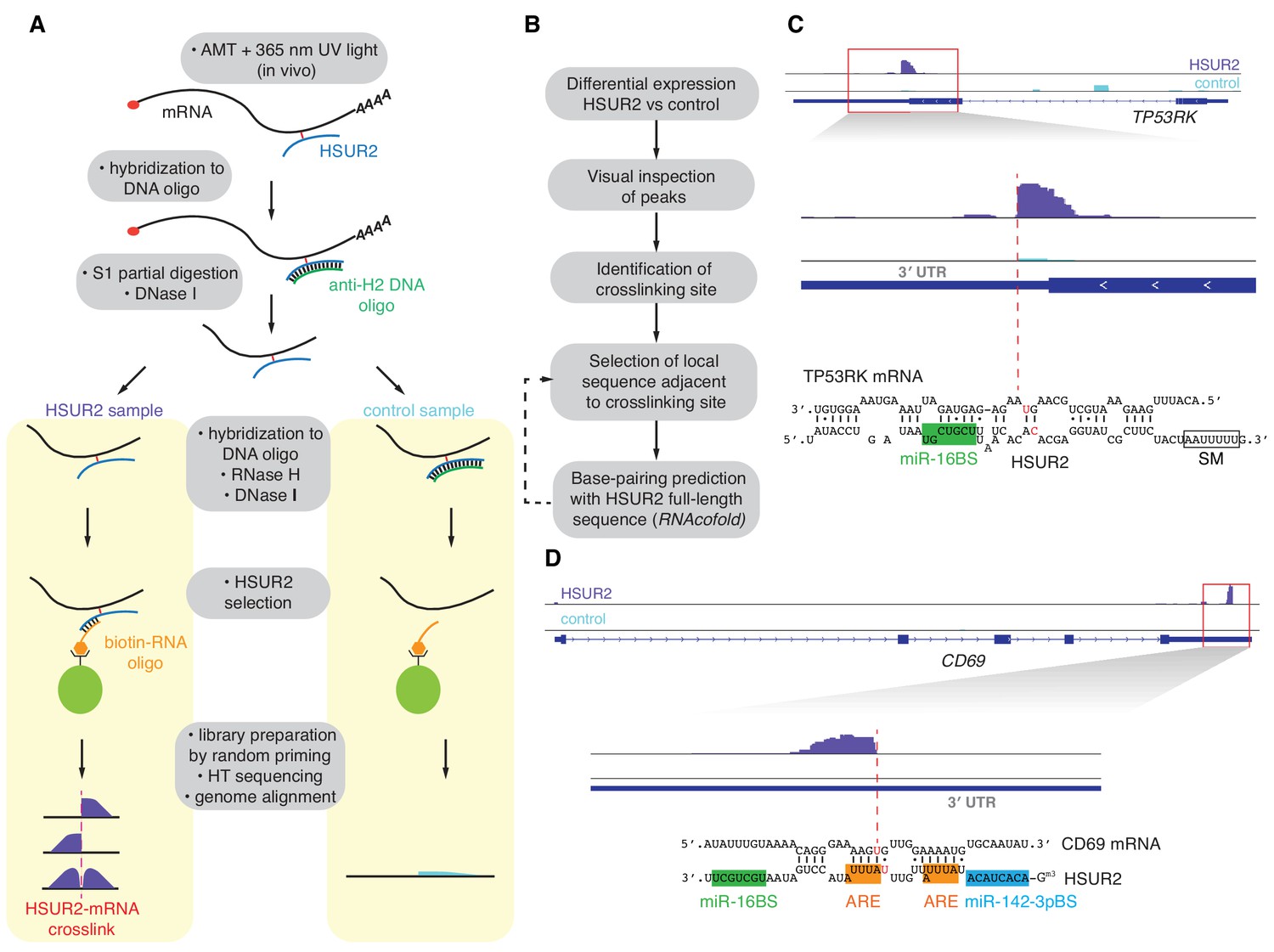
Unambiguous Identification Of Mirna Target Site Interactions By Different Types Of Ligation Reactions Sciencedirect
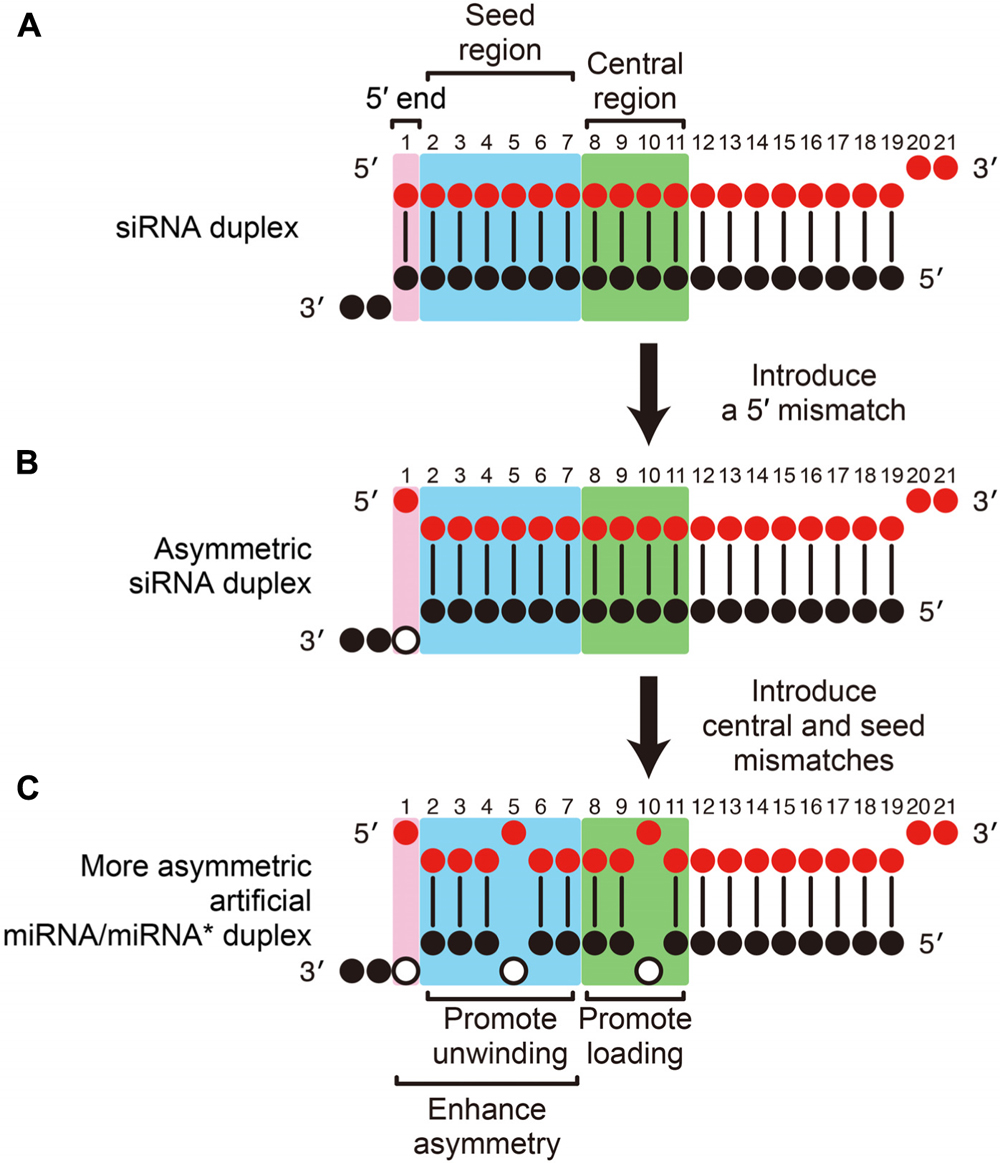



Frontiers Mirna Like Duplexes As Rnai Triggers With Improved Specificity Genetics
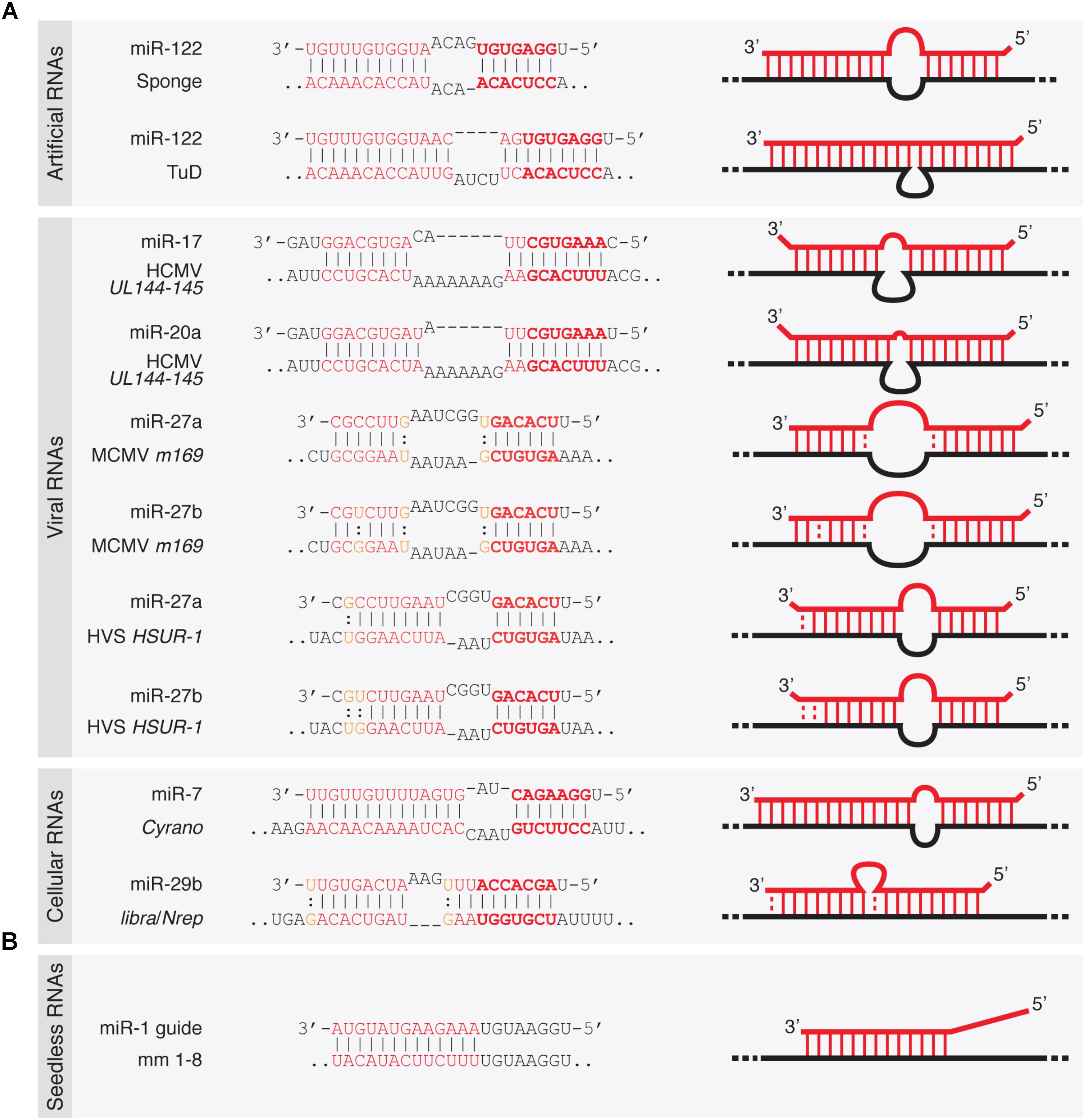



Frontiers Target Rnas Strike Back On Micrornas Genetics
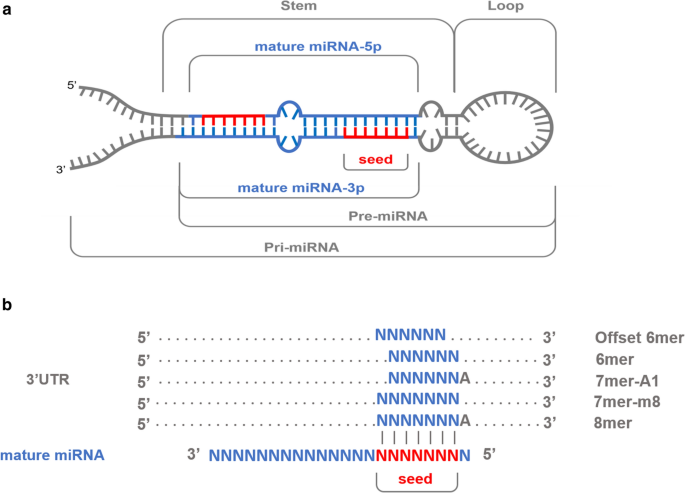



Variability In Porcine Microrna Genes And Its Association With Mrna Expression And Lipid Phenotypes Genetics Selection Evolution Full Text
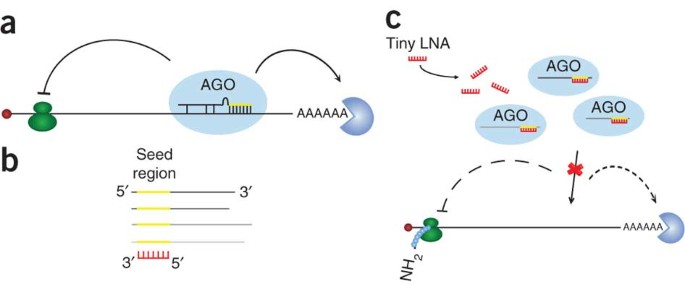



Silencing Of Microrna Families By Seed Targeting Tiny Lnas Nature Genetics




Different Mirna Mrna Seed Site Interaction Patterns 6m Open I




Pdf Microrna Seed Region Length Impact On Target Messenger Rna Expression And Survival In Colorectal Cancer Semantic Scholar



Mirna Workshop Mirna Target Prediction In Animals Ppt Download



Evaluation And Control Of Mirna Like Off Target Repression For Rna Interference Springerlink




Beyond The Seed Structural Basis For Supplementary Microrna Targeting By Human Argonaute2 The Embo Journal



Predicting Effective Microrna Target Sites In Mammalian Mrnas Elife




Mirna Seed Types Nine Seed Types Are Categorized In Two Groups Download Scientific Diagram




Mapping The Human Mirna Interactome By Clash Reveals Frequent Noncanonical Binding Cell



Mirvestigator Framework Detect The Mirnas Driving Co Expression Signatures




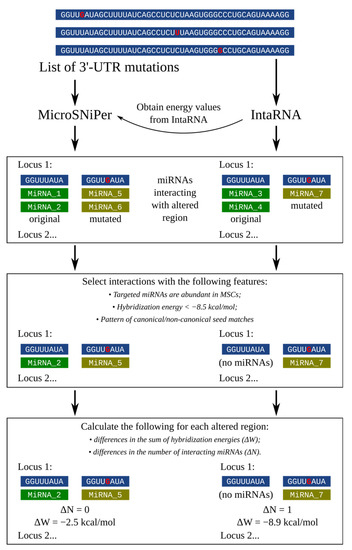
Cimb Free Full Text Evaluating The Effect Of 3 Utr Variants In Dicer1 And Drosha On Their Tissue Specific Expression By Mirna Target Prediction Html



Frontiers Mirna Regulatory Functions In Photoreceptors Cell And Developmental Biology




Types Of Mirna Target Sites And Multiple Sites A Stringent Seed Download Scientific Diagram



1




Mirpro A Novel Standalone Program For Differential Expression And Variation Analysis Of Mirnas Scientific Reports




Mirnas Single Nucleotide Polymorphisms Snps And Age Related Macular Degeneration Amd



Plos Computational Biology Miraw A Deep Learning Based Approach To Predict Microrna Targets By Analyzing Whole Microrna Transcripts



Frontiers Common Features Of Microrna Target Prediction Tools Genetics




Dbmts A Comprehensive Database Of Putative Human Microrna Target Site Snvs And Their Functional Predictions Abstract Europe Pmc



Identifying Microrna Targets In Different Gene Regions Bmc Bioinformatics Full Text




A Study Of Micrornas In Silico And In Vivo Bioimaging Of Microrna Biogenesis And Regulation Kim 09 The Febs Journal Wiley Online Library
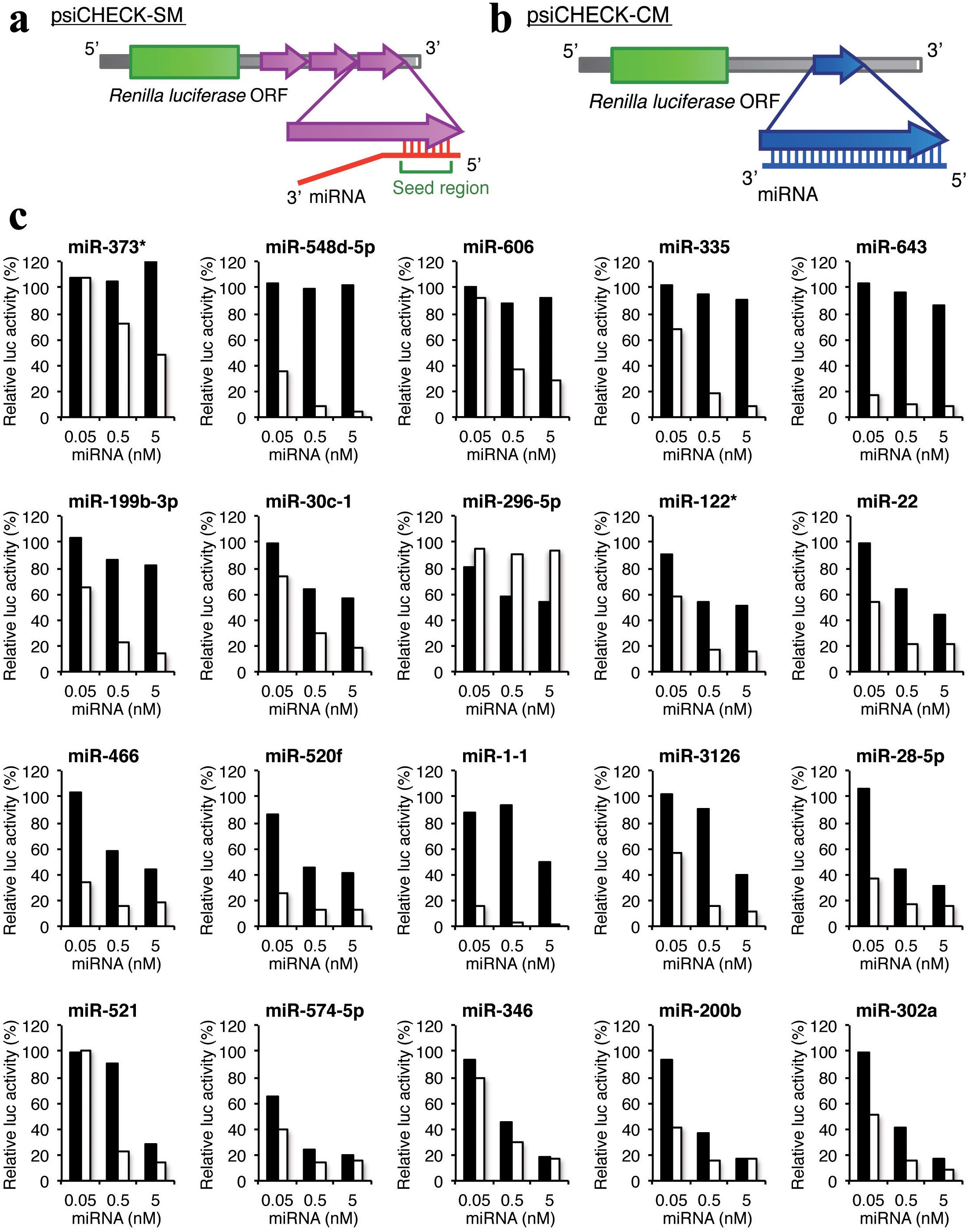



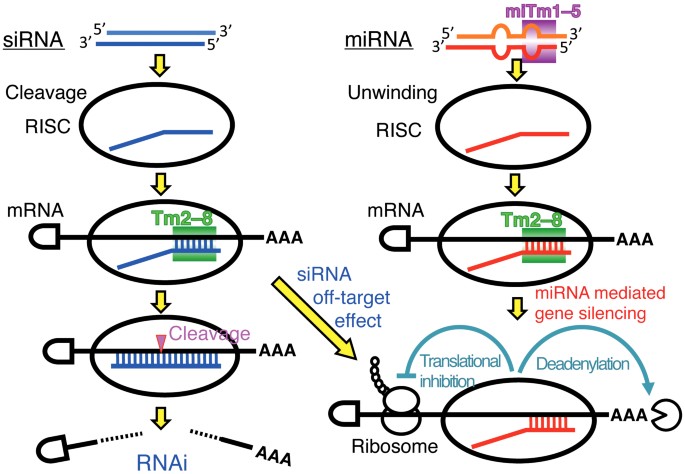
Stability Of Mirna 5 Terminal And Seed Regions Is Correlated With Experimentally Observed Mirna Mediated Silencing Efficacy Scientific Reports



Thegrenze Com




Here S A Topic We Applied Biological Materials Abm Facebook




Different Seed Match Regions Of Mirnas Mirnalyze Follows A Download Scientific Diagram



Identification Of Non Coding Cellular Rna As New Antiviral Targets




3 Uridylation Confers Mirnas With Non Canonical Target Repertoires Sciencedirect




The Biochemical Basis Of Microrna Targeting Efficacy




Examples Of Mirna Target Interactions Pairing Schemes Of Several Download Scientific Diagram



Cs Ucf Edu




Extracellular Micrornas In Human Circulation Are Associated With Mirisc Complexes That Are Accessible To Anti Ago2 Antibody And Can Bind Target Mimic Oligonucleotides Pnas




Human Polymorphism At Micrornas And Microrna Target Sites Pnas




Pairing Beyond The Seed Supports Microrna Targeting Specificity Sciencedirect




Microrna Wikipedia




Crispr Screening Strategies For Microrna Target Identification Yang The Febs Journal Wiley Online Library




Mirna Therapeutics A New Class Of Drugs With Potential Therapeutic Applications In The Heart Future Medicinal Chemistry



Cimb Free Full Text Evaluating The Effect Of 3 Utr Variants In Dicer1 And Drosha On Their Tissue Specific Expression By Mirna Target Prediction Html



Ijms Free Full Text Deciphering Mirnas Action Through Mirna Editing Html




Rna Interference The Molecules That Govern Genetic Control Young Scientists Journal




Mapping The Human Mirna Interactome By Clash Reveals Frequent Noncanonical Binding Cell



Stability Of Mirna 5 Terminal And Seed Regions Is Correlated With Experimentally Observed Mirna Mediated Silencing Efficacy Scientific Reports




Snps In Microrna Target Sites And Their Potential Role In Human Disease Open Biology




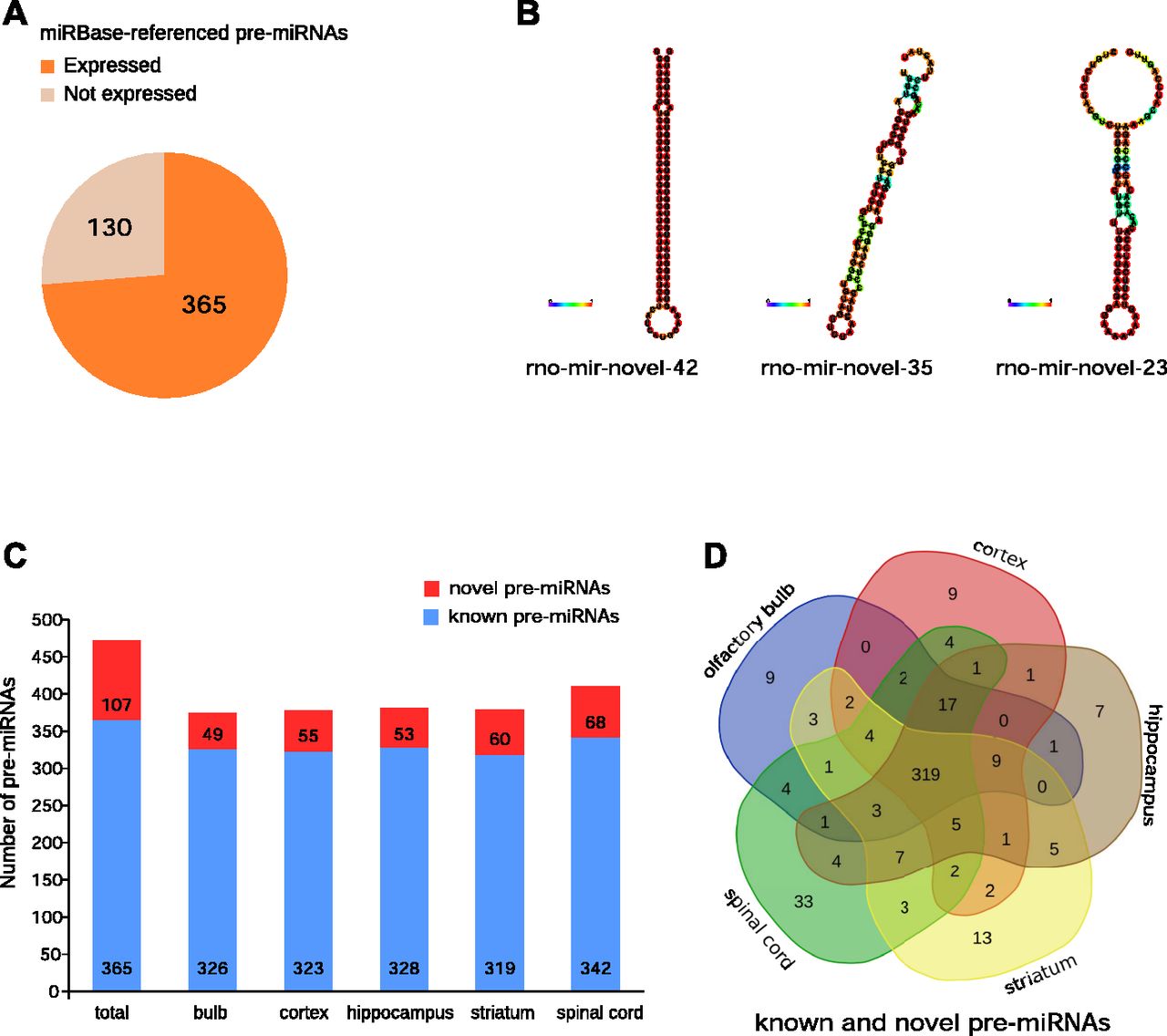
Small Rna Seq Reveals Novel Mirnas Shaping The Transcriptomic Identity Of Rat Brain Structures Life Science Alliance
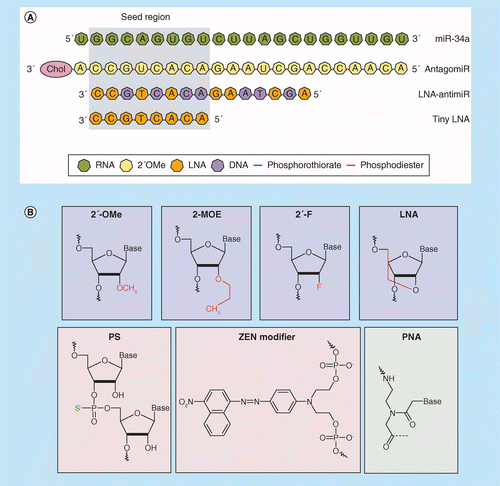



Mirna Therapeutics A New Class Of Drugs With Potential Therapeutic Applications In The Heart Future Medicinal Chemistry
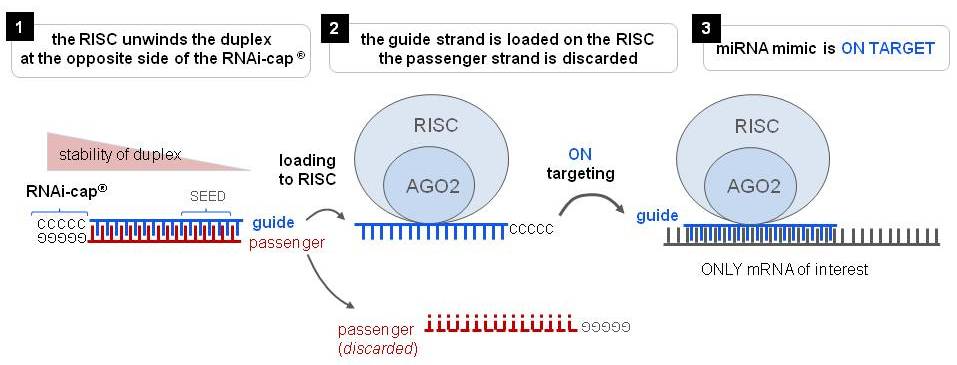



Riboxx Rna Technologies Benefits Of Rnai Cap For Mirna



Mirnasnp V3




Helix 7 In Argonaute2 Shapes The Microrna Seed Region For Rapid Target Recognition The Embo Journal



2




Beyond The Seed Structural Basis For Supplementary Microrna Targeting By Human Argonaute2 The Embo Journal



Microrna Wikipedia




Mirna Targeting Growing Beyond The Seed Trends In Genetics




A Microrna Guide For Clinicians And Basic Scientists Background And Experimental Techniques Heart Lung And Circulation




Microrna Polymorphisms The Future Of Pharmacogenomics Molecular Epidemiology And Individualized Medicine Pharmacogenomics




Figure 1 Micrornas Regulate Key Effector Pathways Of Senescence
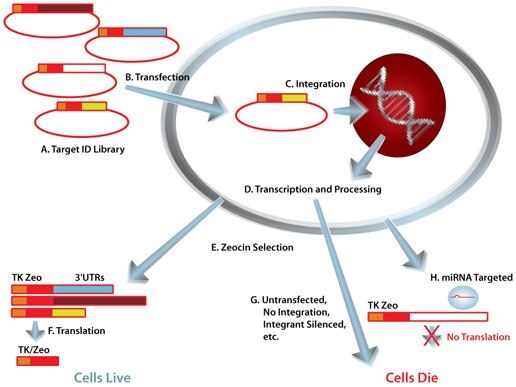



Mission Target Id Library For Human Mirna Target Id And Discovery



2




Clustering Pattern And Functional Effect Of Snps In Human Mirna Seed Regions



2
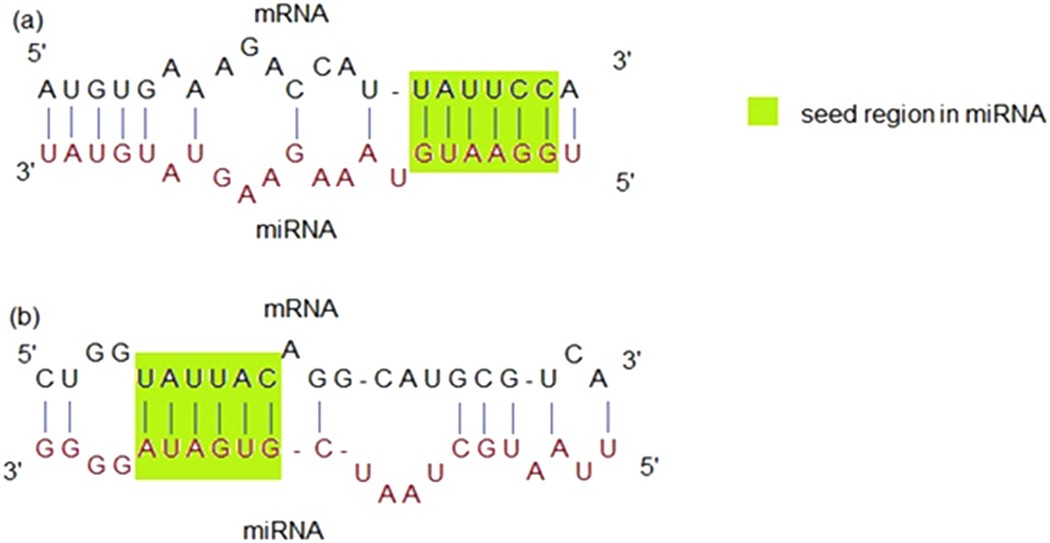



Mirepress Modelling Gene Expression Regulation By Microrna With Non Conventional Binding Sites Scientific Reports



Mirna



Plos One Micrornas Mir 19 Mir 340 Mir 374 And Mir 542 Regulate Mid1 Protein Expression




Structural Differences Between Pri Mirna Paralogs Promote Alternative Drosha Cleavage And Expand Target Repertoires Sciencedirect



1
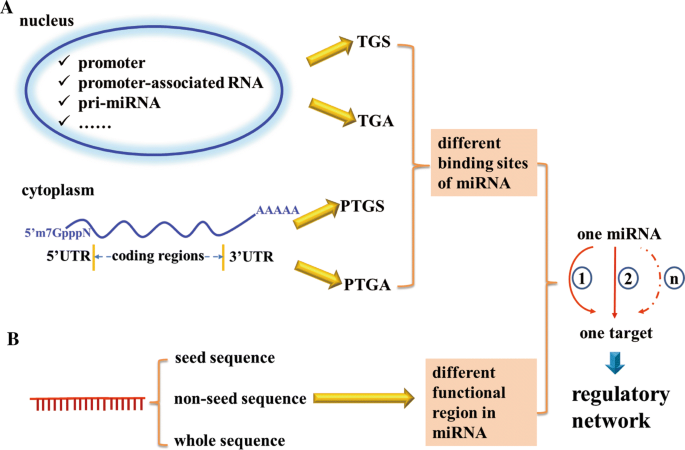



Regulatory Network Of Mirna On Its Target Coordination Between Transcriptional And Post Transcriptional Regulation Of Gene Expression Springerlink




Sirna Versus Mirna As Therapeutics For Gene Silencing Molecular Therapy Nucleic Acids
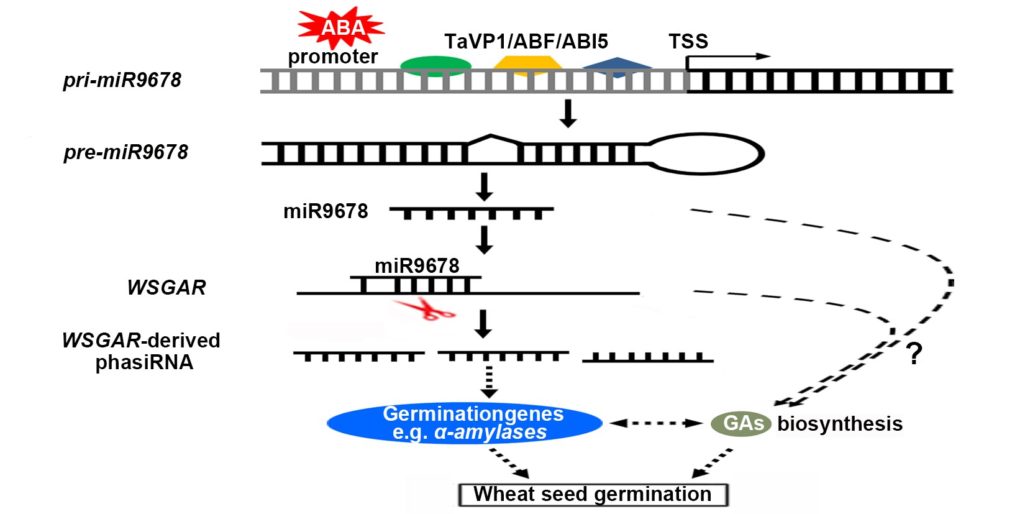



Plantae Mirna Mediated Regulation Of Germination Plantae




Systematic Prediction Of The Impacts Of Mutations In Microrna Seed Sequences



2



1
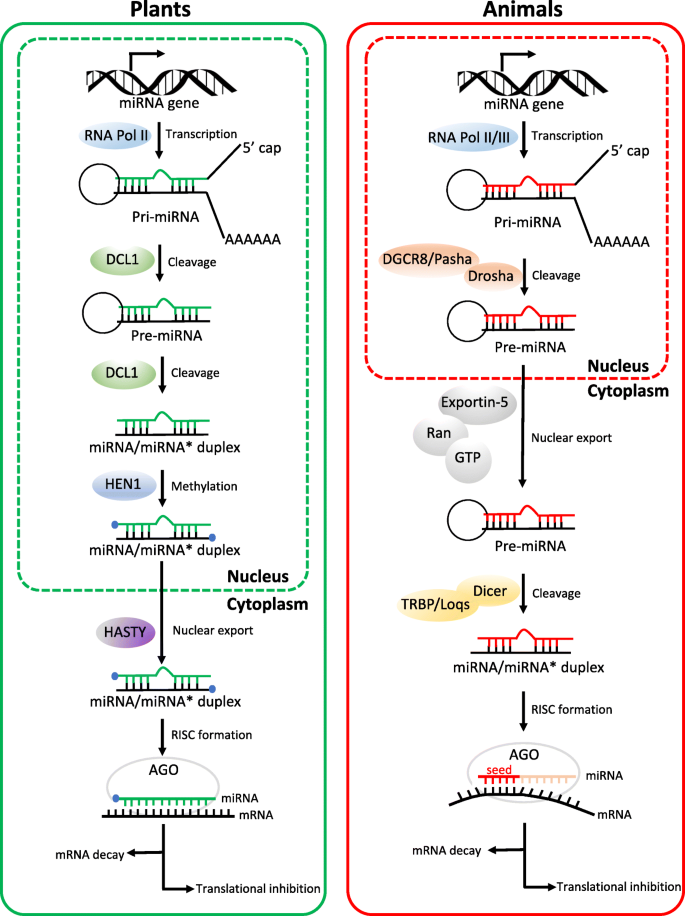



Micrornas From Plants To Animals Do They Define A New Messenger For Communication Nutrition Metabolism Full Text




The Characterization Of Microrna Mediated Gene Regulation As Impacted By Both Target Site Location And Seed Match Type




Sites Matching In The Mirna Seed Region Including All K Mer 8mer Download Scientific Diagram




Microrna Www Gene Quantification Info



2
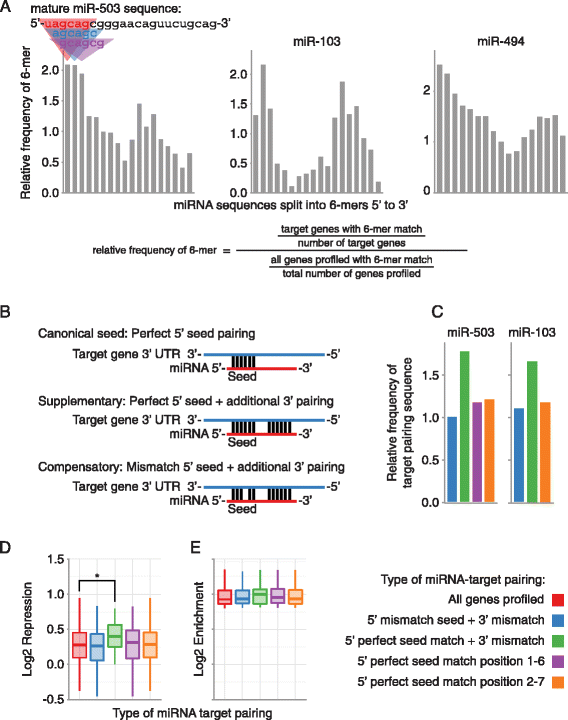



Mir 503 Represses Human Cell Proliferation And Directly Targets The Oncogene Ddhd2 By Non Canonical Target Pairing Bmc Genomics Full Text




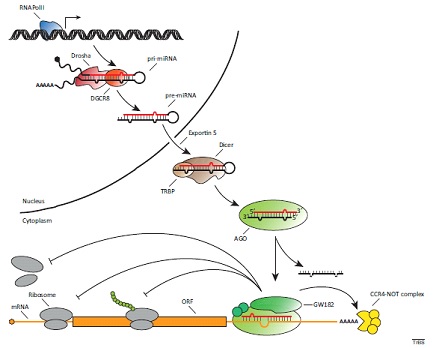
Mirna Introduction Biogenesis Nomenclature And Experimental Workflow




Snps In Microrna Target Sites And Their Potential Role In Human Disease Open Biology




Tools For Sequence Based Mirna Ta Preview Related Info Mendeley



0 件のコメント:
コメントを投稿